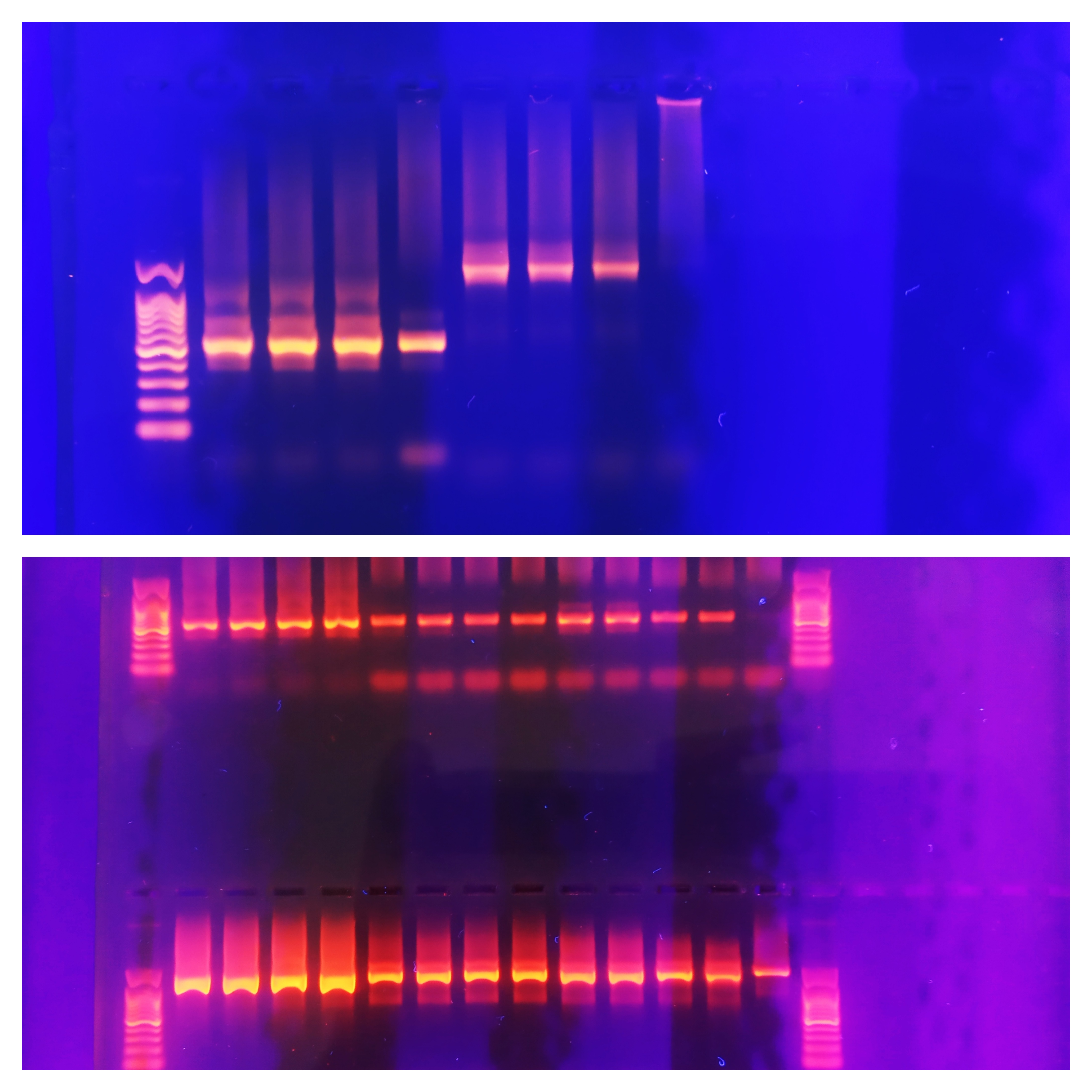
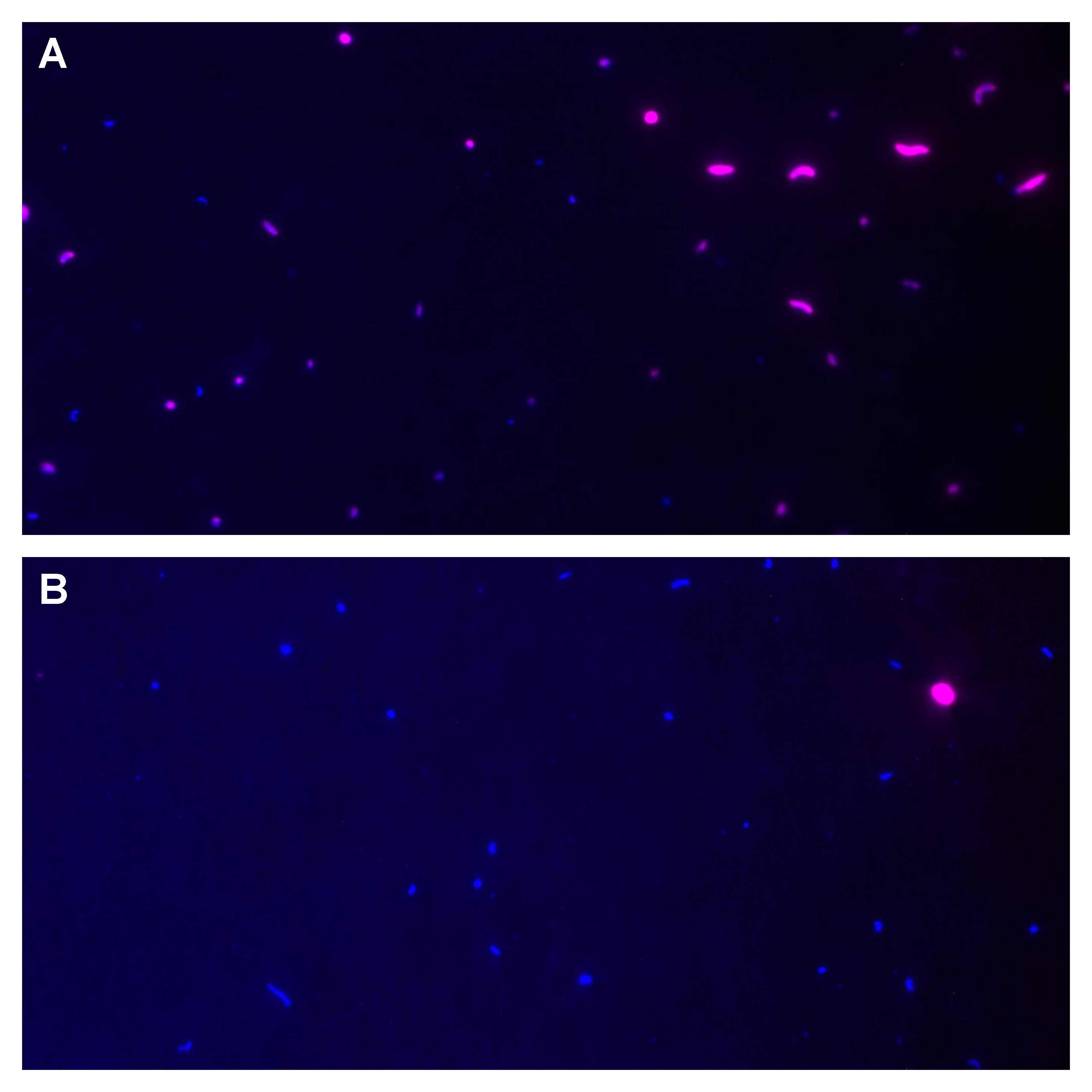
Recent advances in new molecular biology methods and next generation sequencing (NGS) technologies have revolutionized metabarcoding-based research to study complex microbial communities in diverse environments. The inevitable first step in sample preparation is the isolation of DNA, which introduces a number of biases and inconsistencies.In this study, we evaluated the effects of five DNA isolation methods (B1: standard phenol/chloroform/isoamyl extraction, B2 and B3: isopropanol and ethanol precipitation, -both B1 modifications, K1: DNeasy PowerWater Kit (QIAGEN) , K2: a modified DNeasy PowerWater Kit (QIAGEN)) and a direct PCR approach (P), which completely bypasses the isolation step, were evaluated for DNA composition and yield of bacterial “mock-ups” and marine communities from the Adriatic Sea.
Continue reading